Welcome to Molecules@gnu-darwin.org
Molecule News and Molecule of the Day
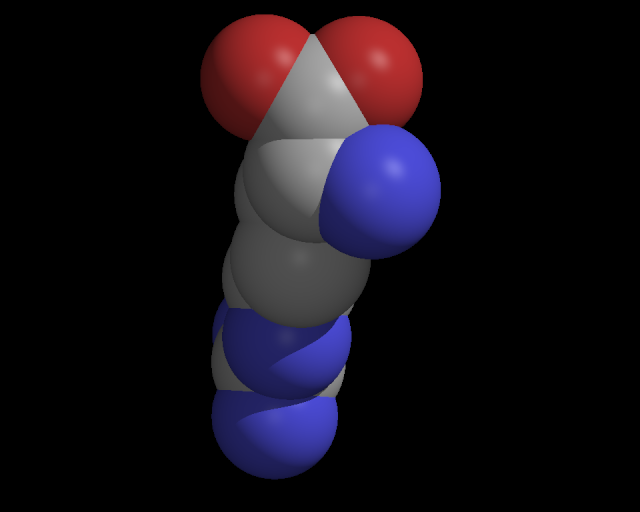
This site is under constant constructionThe molecule of the day is: GDPGDP is the di-phosphate form of guanosine, which is a purine like adenosine. It is one of the hydolized forms of GTP, which bears celluar energy in a manner similar to ATP and other cellular nucleotides. Although GTP is not preferred by most of the energy consumers in the cell, some do prefer it, and the product of that reaction is often GDP. GDP production probably falls far below that of ADP. Guanosine, like adenosine, is a molecule of much interest, and a crucial molecule in many cellular signalling pathways; in metabolism, development, production of proteins, and in the structure of the the gene itself. Light detection proceeds through a guanosine dependent pathway, without which we would be blind. Thus, it appears to be a molecule of some importance. In fact, a little foresight may have prevented the recent drop in the GDP, as it were. Some more notes about this structure: GDP is shown at right in a rather satisfactory 3D conformation. Experts will notice right away that it is not in an energetic minimum conformation, although it is close to a local minimum. The reason for this situation is that like many of our molecules, the one at right was taken from a protein structure, where it was bound in this conformation. Phosphate groups such as these have a particular attraction to the N-terminal ends of alpha helices in proteins, and they sometimes adopt bound conformations that are unlikely to be found in solution. The same is true for the base groups, and the bound orientation of these groups in protein stuctures can be a clue to those who study biochemical mechanisms. Finally, updates to MOD have again been stymied for a few days, due to various reasons, notably election day. Our users may not realize how much of our content is generated by automatic data processing (ADP). If you would like to see how this is done, take a look at the infrastructure, which may be found in the left sidebar. This is how we are able to generate pages like the Vote-more page. GDP related molecules gallery and structural archive SMILES: P(=O)(O)(OCC1OC(C(O)C1O)N2C=NC3=C2N=C(N)NC3=O)OP(=O)(O)O other names: 146-91-8 C00035 GDP Guanosine 5'-diphosphate Guanosine diphosphate
related molecule names: |
   pdb file:584548.pdb directory:584548 CAS: 146-91-8
sirius yellow cross-eyed 3D
|
Molecule News
updated Wed Sep 17 23:56:32 EDT 2008Added Molscript graphics and MOD desktop image to the right sidebar. The theme is infinite carbon chain. The code is in infrastructure. The other main molecular graphics programs in use at this time are RasMol, Rester3D, PyMOL, and PovChem. The viewer is xzgv, and ImageMagick for touch up; mainly mogrify and convert. Please feel free to send any questions along.
updated Thu Sep 11 22:33:51 EDT 2008
We continue to get encouragment to provide coordinate files with improved geometry, although this is a computationally intensive task which is down the road still. Meanwhile, a few supurb 3-dimensional structures are already found in the archive, and two of them have been featured in the right sidebar today. If you rummage around in the MOD galleries, you are likely to find more 3D coordinate files, as well as some very interesting 2D representations! In our opinion, many of the most interesting molecules have planar geometry, so that the current files are quite useful. In due course, we plan to provide a comprehensive collection of coordinate files with good stereochemical geometry, all in the selfsame spirit of free and open source software and public access. We welcome any and all input.
updated Wed Sep 10 20:28:49 EDT 2008
Our new directory style was featured in a the right sidebar of the Jasmone MOD page today. Just click the directory link from the top page or in any of the recent galleries to view this new feature. These pages typically include links to the source databases, which often have chemical diagrams or useful additional information about the molecules. A smiley code is currently featured at the bottom of each page. Smile.
updated Thu Sep 4 22:28:57 EDT 2008
Added quick navigation links to the left side bar. The site has reached 111 GB, and a massive backup is underway. We are still just getting started.
updated Thu Sep 4 13:30:20 EDT 2008
Added developer links to the left side bar.
updated Wed Sep 3 12:05:13 EDT 2008
I have automated the process to make molecule galleries, which I am finding very useful to my research, so I thought that I would share the opportunity and take requests. It is quick and easy. I did a bisphenol search today, and as I expected, the search turned up a few stilbenes, which is the same molecular family as resveratrol. Strikingly, Bisphenol A is a noted teratogen, which shows that care must be taken when using such molecules during pregnancy, nursing, and development. Here is a link to the bisphenol gallery.
http://molecules.gnu-darwin.org/mod/Bisphenol-more.html
If you would find it useful, I am definitely interested in taking requests for molecule galleries. Just send me the name of your favorite molecule. As you may be able to tell, we have access for galleries to about 1% or 0.5 million structures, in the total archive thus far, but it is already quite useful. The automated jobs are underway to fill in the rest. The next milestone is the capability to index and make galleries for the remaining millions of molecules for which we have pdb files already, which would be 5-10% complete. Any comments or suggestions are also welcome.
updated Tue Sep 2 14:10:15 EDT 2008
A terminal screenshot has been added to the right sidebar, in order to show the kind of code that is being used to generate the Molecules directory indices. All inquiries and suggestions are welcome.
updated Fri Aug 29 14:39:14 EDT 2008
Reaction to our website has been quite positive and encouraging, but people are anxious to see structures with improved stereochemistry. We have received many suggestions about how to make that happen faster. For the time being, we are experiencing the best structure viewing results with RasMOL. Name extraction and structure file linking jobs are underway, which is an indication that we are still in a preliminary phase. Google spiders more almost everyday, and past experience indicates that the search capability will become very useful indeed! Here is some secret mumbo jumbo. I'm afraid that my Japanese or Korean is broken at best ;-}. Enjoy!
updated Wed Aug 27 21:38:32 EDT 2008
An RSS News Feed has been added to the right side bar.
updated Tue Aug 26 19:27:29 EDT 2008
Several new indices have been added to our sidebar, notably; current index by name, and gallery by name. These are large indices, but also vastly incomplete, and work is underway to improve them. A list of bloggers who have mentioned the site, with links in their blogs, is now in the front sidebar, although only one is known thus far. Due to the vastness of the scale, many of these things will be divided out and buried a little deeper eventually, but they can be showcased now, and they are a good survey.
updated Tue Aug 19 19:45:09 EDT 2008
Some facts: The Molecules website contains more than 4 million small molecule structure files in pdb format, and molecular graphics representations. About 50 million molecules are still in the pipe, and they are expected to appear here over the course of the next few weeks and months. The pdb format is readable by common FOSS molecule viewer software, such as RasMol and PyMOL. In due course, we plan to provide high quality structures via energy minimization refinement, and additional resources.
Molecules@gnu-darwin.org is founded in the spirit of free software, open source, and public access. It is hoped that access to these files will be a wonderful community resource for science education, research, and entertainment as well. We are looking for investment, funding, or sponsorship in order to expedite and expand this work, and lead the field, with an eye towards an advanced, complete, synthetic, structural, and informatical bioorganome. Meanwhile, the site is already an exceptional lab resource, and molecular catalog, providing the means and building blocks towards additional novel structures. We aim to be the best.
The structural biology, protein crystallography, and molecular graphics talent that is building the Molecules archive is available to work for you in a contract or consulting arrangement. Wide-ranging expertise is available. Molecules@gnu-darwin.org is built entirely with FOSS, free and open source software, GNU-Darwin OS, and it is under the aegis of The GNU-Darwin Distribution. Here is a link to the Distribution résumé. Our founder is an X-ray laboratory admin for the Department of Biophysics and Biophysical Chemistry of Johns Hopkins University School of Medicine. You can also read his CV. We would like to build a community around this website, and we are looking for volunteers and collaborators to help. Regarding any aspect of the work of this site, please feel free to contact us, molecules@gnu-darwin.org, with gdmolecules in the subject line. Cheers!