
|
Welcome to Molecules@gnu-darwin.org
Molecule News and Molecule of the Day
|
|
This site is under constant construction
The molecule of the day is: ATP
ATP, Adenosine 5'-triphosphate, is known as the energy molecule of the cell,
due to the fact that the cell stores much of its energy in second
phosphodiester bond, which is the di-phoshate bond furthest to the left in
the figure shown at right. Proteins are able to release the energy in this
bond in order to do the work of the cell, and the coordination of a
magnesium ion is required to do this. Adenosine receptor is a noted
target of caffeine, which is similar in structure to adenosine, and it
probably affects other adenosine-related activities as well.
In addition to the key role that adenosine plays in cellular energy
metabolism, it plays many other crucial roles in the
cell, such as; in signalling pathways, and as part of the fundamental
structure of RNA and DNA (in the deoxy form). It is literally part of the
the stucture that encodes the genetic secrets of life, which are now
beginning to be known in great detail, by which we can begin to increase
longevity. Adenosine is also part of the signal
by which the cell recognizes when to replicate its genes, when to make
protein, when to propogate, but also when not to, when to live,
and when to die. This purine class molecule is truly gate and gate keeper.
ATP related molecules gallery and structural archive
other names:
56-65-5 ATP Adenosine 5'-triphosphate C00002
Canonical SMILES:
C1=NC2=C(C(=N1)N)N=CN2C3C(C(C(O3)COP(=O)(O)OP(=O)(O)OP(=O)(O)O)O)O
related molecule names:
58337-43-2 Adenosine 5'-tetraphosphate CPD-607 adenosine tetraphosphate
56-65-5 ATP Adenosine 5'-triphosphate C00002
20398-34-9 ADP Adenosine 5'-diphosphate C00008
5'-AMP 5'-Adenosine monophosphate 5'-Adenylic acid 61-19-8 AMP Adenosine 5'-monophosphate Adenosine 5'-phosphate Adenylate Adenylic acid C00020
1053-73-2 3'-Phosphoadenylate Adenosine 3',5'-bisphosphate C00054 PAP Phosphoadenosine phosphate
58-61-7 Adenosine C00212
485-84-7 5'-Adenylyl sulfate APS Adenosine 5'-phosphosulfate Adenylyl sulfate C00224
ADP-glucose Adenosine diphosphoglucose C00498
3',5'-Cyclic AMP 60-92-4 Adenosine 3',5'-phosphate C00575 Cyclic AMP Cyclic adenylic acid cAMP
130-49-4 2'-Adenylic acid AMP 2'-phosphate Adenosine 2'-phosphate Adenosine-2'-monophosphate C00946
3'-AMP 3'-Adenosine monophosphate 3'-Adenylic acid 84-21-9 AMP 3'-phosphate Adenosine 3'-phosphate Adenosine-3'-monophosphate C01367
Adenosine 5'-tetraphosphate Adenosine tetraphosphate C03483
3805-37-6 Adenosine 2',5'-bisphosphate C03850
Adenosine 5'-phosphoramidate C03851
Adenosine 5'-(gamma-thio)triphosphate C04380
Adenosine-5'-phosphoselenate Adenylylselenate C05686
3',5'-Adenosine 5'-diphosphate C05709
Adenosine-GDP-cobinamide Adenosylcobinamide-GDP C06510
3'-Amino-3'-deoxy-N6,N6-dimethyladenosine 58-60-6 6-Dimethylamino-9-(3'-ribosylamine)purine ARDMA Adenosine, 3'-amino-3'-deoxy-N,N-dimethyl- Aminonucleoside Aminonucleoside puromycin NSC3056 PUROMYCIN AMINONUCLEOSIDE Puromycin, aminonucleoside SAN Stylomycin aminonucleoside
2,6-Diaminonebularine 2,6-Diaminopurine ribonucleoside 2,6-Diaminopurine riboside 2,6-Diaminopurinosine 2-Aminoadenosine 2096-10-8 9-.beta.-Ribosyl-2,6-diaminopurine 9H-Purine, 2,6-diamino-9-.beta.-D-ribofuranosyl- 9H-Purine-2,6-diamine, 9-.beta.-D-ribofuranosyl- Adenosine, 2-amino- NSC7363
.beta.-Adenosine .beta.-D-Adenosine .beta.-D-Ribofuranose, 1-(6-amino-9H-purin-9-yl)-1-deoxy- .beta.-D-Ribofuranoside, adenine-9 58-61-7 6-Amino-9.beta.-D-ribofuranosyl-9H-purine 9.beta.-D-Ribofuranosyladenine 9H-Purin-6-amine, 9.beta.-D-ribofuranosyl- ADENOSINE Adenine nucleoside Adenine riboside Adenosin Boniton Myocol NSC7652 Nucleocardyl Sandesin Usaf cb-10 WLN: T56 BN DN FN HNJ IZ D- BT5OTJ CQ DQ E1Q
1818-71-9 2-Hydroxyadenosine Adenosine, 1,2-dihydro-2-oxo- Crotonosid Crotonoside Isoguanine riboside Isoguanine, 9-.beta.-D-ribofuranosyl- Isoguanosine NSC12161
18048-85-6 Adenosine, 5'-O-trityl- NSC14167
2504-55-4 3'-Amino-3'-deoxyadenosine 9H-Purine, 6-amino-9-(3-amino-3-deoxy-.beta.-D-ribofuranosyl)- ADENOSINE,-3'-AMINO-3'-DEOXY Adenosine, 3'-amino-3'-deoxy- NSC18192 Spalgomycin
5'-AMP 5'-Adenylic acid 61-19-8 9H-Purine, 6-amino-9-.beta.-D-ribofuranosyl-, 5'-phosphate A 5MP ADENYLIC ACID, YEAST AMP AMP (nucleotide) Adenosine 5'-(dihydrogen phosphate) Adenosine 5'-monophosphate Adenosine 5'-monophosphoric acid Adenosine 5'-phosphate Adenosine 5'-phosphoric acid Adenosine Phosphate Adenosine monophosphate Adenosine, mono(dihydrogen phosphate) (ester) Adenovite Adenylic acid Cardiomone Lycedan Muscle adenylic acid My-B-Den NSC20264 Phosaden Phosphentaside
1867-73-8 6-Methyladenosine 6-Methylaminopurine D-riboside 6-Methylaminopurine ribonucleoside 6-Methylaminopurine riboside 6-Methylaminopurinosine Adenosine, N-methyl- N-Methyladenosine N6-Methyladenosine NSC 29409 NSC29409
2',3'-Isopropylideneadenosine 2',3'-O-Isopropylideneadenosine 362-75-4 Adenosine, 2',3'-O-(1-methylethylidene)- Adenosine, 2',3'-O-isopropylidene- NSC29413
146-78-1 2-FAR 2-FAS 2-Fluoroadenosine Adenosine, 2-fluoro- NSC30605 Purine, 6-amino-2-fluoro-9-.beta.-D-ribofuranosyl- SRI 727
146-77-0 2 ClAdo 2-Chloroadenosine Adenosine, 2-chloro- Cl AS NSC36896
2-Methoxyadenosine 24723-77-1 Adenosine, 2-methoxy- NSC36899
4-Amino-7-.beta.-D-ribofuranosyl-7H-pyrrolo[2,3-d]pyrimidine 4-Amino-7.beta.-D-ribofuranosyl-7H-pyrrolo(2,3-d)pyrimidine 69-33-0 7-.beta.-D-ribofuranosyl-7H-pyrrolo-[2,3-d]pyrimidin-4-amine 7-Deazaadenosine 7-Deazadenosine 7.beta.-D-Ribofuranosyl-7H-pyrrolo(2,3-d)pyrimidine-4-amine 7H-Pyrrolo(2,3-d)pyrimidine, 4-amino-7.beta.-D-ribofuranosyl- 7H-Pyrrolo[2,3-d]pyrimidin-4-amine, 7-.beta.-D-ribofuranosyl- 7H-Pyrrolo[2,3-d]pyrimidine, 4-amino-7-.beta.-D-ribofuranosyl- Adenosine, 7-deaza- Antibiotic 155b2t B 120121 NSC 56408 NSC-56408 NSC56408 SKI 26996 Sparsomycin A TBC TUBERCIDIN Tubercidine U 10071 U10071 WLN: T56 BN GN INJ FZ I- BT5OTJ CQ DQ E1Q
.beta.-D-erythro-Pentofuranoside, adenine-9 3-deoxy- 3'-Deoxyadenosine 3-DEOXYADENOSINE 73-03-0 9-(.beta.-D-3'-Deoxyribofuranosyl)adenine 9-Cordyceposidoadenine 9H-Purin-6-amine, 9-(3-deoxy-.beta.-D-erythro-pentofuranosyl)- 9H-Purine, 6-amino-9-(3-deoxy-.beta.-D-ribofuranosyl)- Adenosine, 3'-deoxy- CORDYCEPIN Cordycepine NSC63984
4294-16-0 6-Benzyladenosine 6-Benzylamino-9.beta.-D-ribofuranosylpurine 6-Benzylamino-9.beta.-ribofuranosylpurine, D- 6-Benzylaminopurine riboside Adenosine, N-(phenylmethyl)- Adenosine, N-benzyl- Benzoadenosine Benzyladenine ribonucleoside Benzyladenine riboside Benzyladenosine Benzylaminopurine riboside N-6-Benzyladenosine N-Benzyladenosine N6-Benzyladenosine N6BAR NSC70423 Pyranylbenzyladenine
2'-Deoxy-5'-adenylic acid 2'-Deoxy-AMP 2'-Deoxyadenosine 5'-monophosphate 2'-Deoxyadenosine 5'-phosphate 2'-Deoxyadenosine monophosphate 2'-Deoxyadenylic acid 2'-dAMP 5'-Adenylic acid, 2'-deoxy- 653-63-4 Adenosine, 2'-deoxy-, 5'-(dihydrogen phosphate) Adenosine, 2'-deoxy-, 5'-phosphoric acid Deoxy-5'-adenylic acid Deoxy-AMP Deoxyadenosine 5'-monophosphate Deoxyadenosine 5'-phosphate Deoxyadenosine monophosphate Deoxyadenylic acid NSC77680 PdA dAMP
2946-39-6 8-Bromoadenosine Adenosine, 8-bromo- Bromoadenosine NSC79213
2500-80-3 Adenosine, 4'-thio- NSC82219
2'-Deoxy-5'-adenylic acid 2'-Deoxy-AMP 2'-Deoxyadenosine 5'-monophosphate 2'-Deoxyadenosine 5'-phosphate 2'-Deoxyadenosine monophosphate 2'-Deoxyadenylic acid 2'-dAMP 5'-Adenylic acid, 2'-deoxy- 653-63-4 Adenosine, 2'-deoxy-, 5'-(dihydrogen phosphate) Adenosine, 2'-deoxy-, 5'-phosphoric acid Deoxy-5'-adenylic acid Deoxy-AMP Deoxyadenosine 5'-monophosphate Deoxyadenosine 5'-phosphate Deoxyadenosine monophosphate Deoxyadenylic acid NSC82625 PdA dAMP
.beta.-D-Ribofuranose, 1-(6-amino-9H-purin-9-yl)-1,2-dideoxy- .beta.-D-erythro-Pentofuranoside, adenine-9 2-deoxy- 2'-Deoxyadenosine 2-Deoxyadenosine 958-09-8 9H-Purin-6-amine, 9-(2-deoxy-.beta.-D-erythro-pentofuranosyl)- 9H-Purin-6-amine, 9-(2-deoxy-.beta.-D-ribofuranosyl)- Adenine deoxy nucleoside Adenine deoxyribonucleoside Adenine deoxyribose Adenosine, 2'-deoxy- Adenyldeoxyriboside Deoxyadenosine Desoxyadenosine NSC83258
3'-AMP 3'-Adenosine monophosphate 3'-Adenylic acid 84-21-9 Adenosine 3'-monophosphate Adenosine 3'-phosphate Adenosine-3'-monophosphoric acid Adenosine-3'-phosphate NSC210570 Synadenylic acid Yeast adenylic acid
14365-44-7 5'-Amino-5'-deoxyadenosine 5'-Deoxy-5'-aminoadenosine Adenosine, 5'-amino-5'-deoxy- NSC238990
|

pdb file:3251.pdb
directory:3251
CAS: 56-65-5
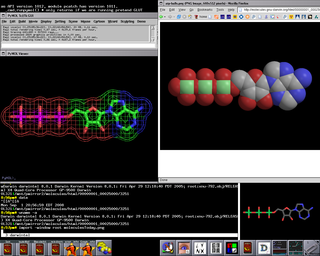
MOD screenshot

MOD termshot

RSS News Feed



(syndicate freely:)
|
Molecule News
updated Tue Sep 2 14:10:15 EDT 2008
A terminal screenshot has been added to the right sidebar,
in order to show the kind
of code that is being used to generate the Molecules directory indices. All
inquiries and suggestions are welcome.
updated Fri Aug 29 14:39:14 EDT 2008
Reaction to our website has been quite positive and encouraging, but people are anxious to
see structures with improved stereochemistry. We have received many suggestions about how
to make that happen faster. For the time being, we are experiencing the
best structure viewing results with RasMOL. Name extraction and structure file linking
jobs are underway, which is an indication that we are still in a preliminary phase. Google
spiders more almost everyday, and past experience indicates that the search capability will
become very useful indeed! Here is some secret mumbo jumbo. I'm afraid that my Japanese
or Korean is broken at best ;-}. Enjoy!
updated Wed Aug 27 21:38:32 EDT 2008
An RSS News Feed has been added to the right side bar.
updated Tue Aug 26 19:27:29 EDT 2008
Several new indices have been added to our sidebar, notably;
current index by name, and
gallery by name. These are large indices, but also
vastly incomplete, and work is underway to improve them.
A list of bloggers who have mentioned the site, with links in their blogs, is now in the front sidebar,
although only one is known thus far. Due to the vastness of the scale,
many of these things will be divided out and buried a little deeper
eventually, but they can be showcased now, and they are a good survey.
updated Tue Aug 19 19:45:09 EDT 2008
Some facts: The Molecules website contains more than
4 million small molecule structure files in pdb format,
and molecular graphics representations. About 50
million molecules are still in the pipe, and they are expected
to appear here over the course of the next few weeks and months.
The pdb format is readable by common FOSS molecule viewer software,
such as RasMol and PyMOL.
In due course, we plan to provide high quality structures via
energy minimization refinement, and additional resources.
Molecules@gnu-darwin.org is founded
in the spirit of free software, open source, and public access.
It is hoped that access to these files will be a wonderful community
resource for science education, research, and entertainment as well.
We are looking for investment, funding, or sponsorship in order to expedite and expand this work, and lead the field,
with an eye towards an advanced, complete, synthetic, structural, and informatical bioorganome.
Meanwhile, the site is already an exceptional lab resource,
and molecular catalog, providing the means and building blocks
towards additional novel structures. We aim to be the best.
The structural biology, protein crystallography, and molecular graphics talent that is
building the Molecules archive is available to work for you in
a contract or consulting arrangement. Wide-ranging expertise is available.
Molecules@gnu-darwin.org is built entirely with
FOSS,
free and open source
software, GNU-Darwin OS, and it is under
the aegis of
The GNU-Darwin Distribution.
Here is a link to the
Distribution
résumé.
Our founder is an X-ray laboratory admin for the
Department of Biophysics and Biophysical
Chemistry of Johns Hopkins University School of Medicine. You can also
read his CV.
We would like to build a community around this website, and we are looking for
volunteers and collaborators to help. Regarding any aspect of the work of this site, please feel free
to contact us,
molecules@gnu-darwin.org, with gdmolecules in the subject line. Cheers!
|






